SDePER
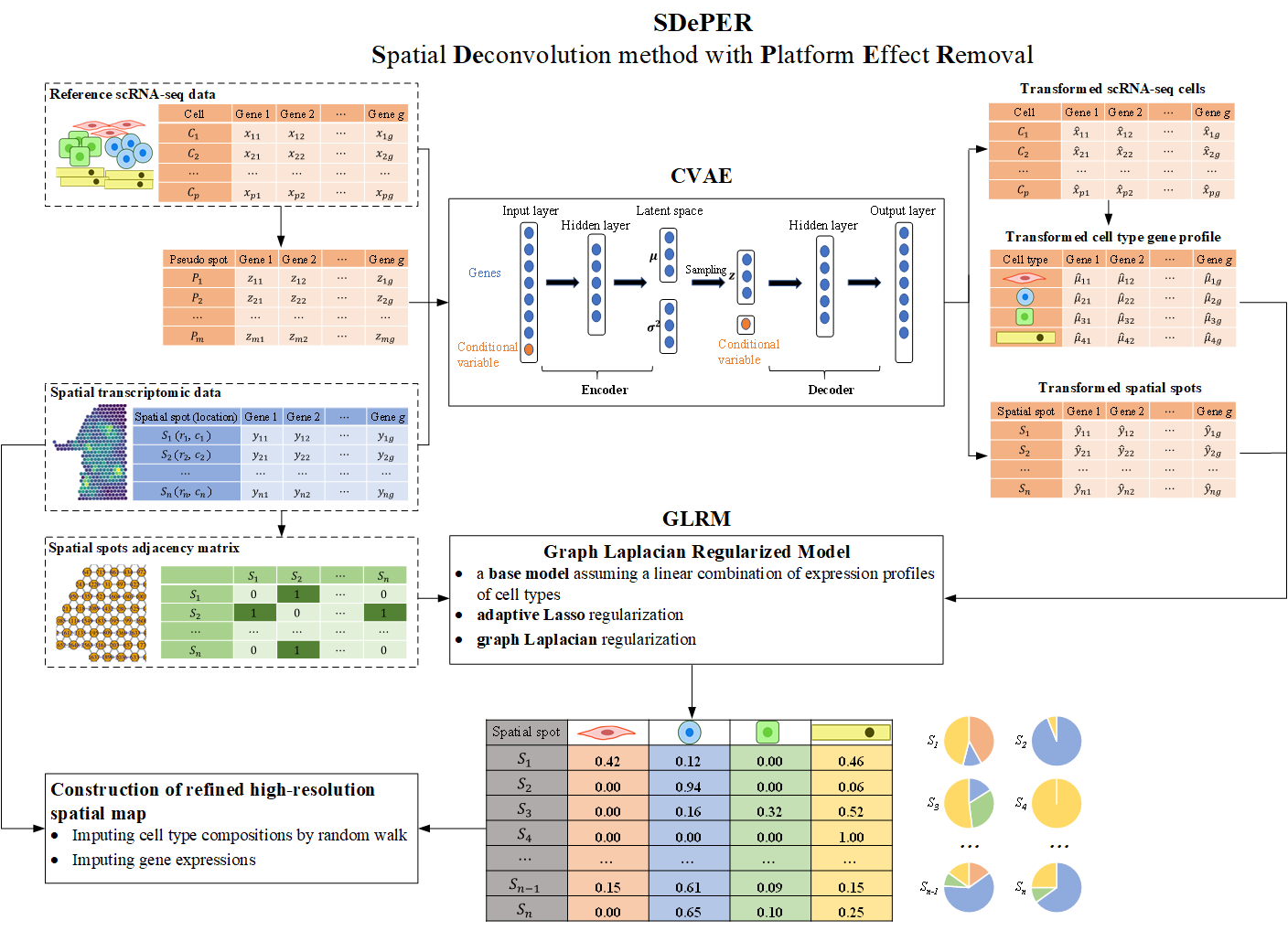
SDePER (Spatial Deconvolution method with Platform Effect Removal) is a hybrid machine learning and regression method to deconvolve Spatial barcoding-based transcriptomic data using reference single-cell RNA sequencing data, considering platform effects removal, sparsity of cell types per capture spot and across-spots spatial correlation in cell type compositions. SDePER is also able to impute cell type compositions and gene expression at unmeasured locations in a tissue map with enhanced resolution.
Quick Start
SDePER currently supports only Linux operating systems such as Ubuntu, and is compatible with Python 3.9.x and 3.10.x releases (3.11+ not yet supported).
SDePER can be installed via conda
conda create -n sdeper-env -c bioconda -c conda-forge python=3.9.12 sdeper
or pip
conda create -n sdeper-env python=3.9.12
conda activate sdeper-env
pip install sdeper
SDePER supports an out-of-the-box feature, meaning that users only need to provide the required four input files for cell type deconvolution. The package manages all aspects of file reading, preprocessing, cell type-specific marker gene identification, and more internally. The required files are:
- raw nUMI counts of spatial transcriptomics data (spots × genes):
spatial.csv - raw nUMI counts of reference scRNA-seq data (cells × genes):
scrna_ref.csv - cell type annotations for all cells in scRNA-seq data (cells × 1):
scrna_anno.csv - adjacency matrix of spots in spatial transcriptomics data (spots × spots; optional):
adjacency.csv
To start cell type deconvolution using all default settings by running
runDeconvolution -q spatial.csv -r scrna_ref.csv -c scrna_anno.csv -a adjacency.csv
Homepage: https://az7jh2.github.io/SDePER/.
Full Documentation for SDePER is available here.
Example data and Analysis using SDePER are summarized in this page. All related materials can be found in the Analysis repository.
Citation
If you use SDePER, please cite:
Yunqing Liu, Ningshan Li, Ji Qi et al. SDePER: a hybrid machine learning and regression method for cell-type deconvolution of spatial barcoding-based transcriptomic data. Genome Biology 25, 271 (2024). https://doi.org/10.1186/s13059-024-03416-2






